
Cheat sheet: Inputs/outputs for user-defined functions
Source:vignettes/cheat-sheet.Rmd
cheat-sheet.Rmd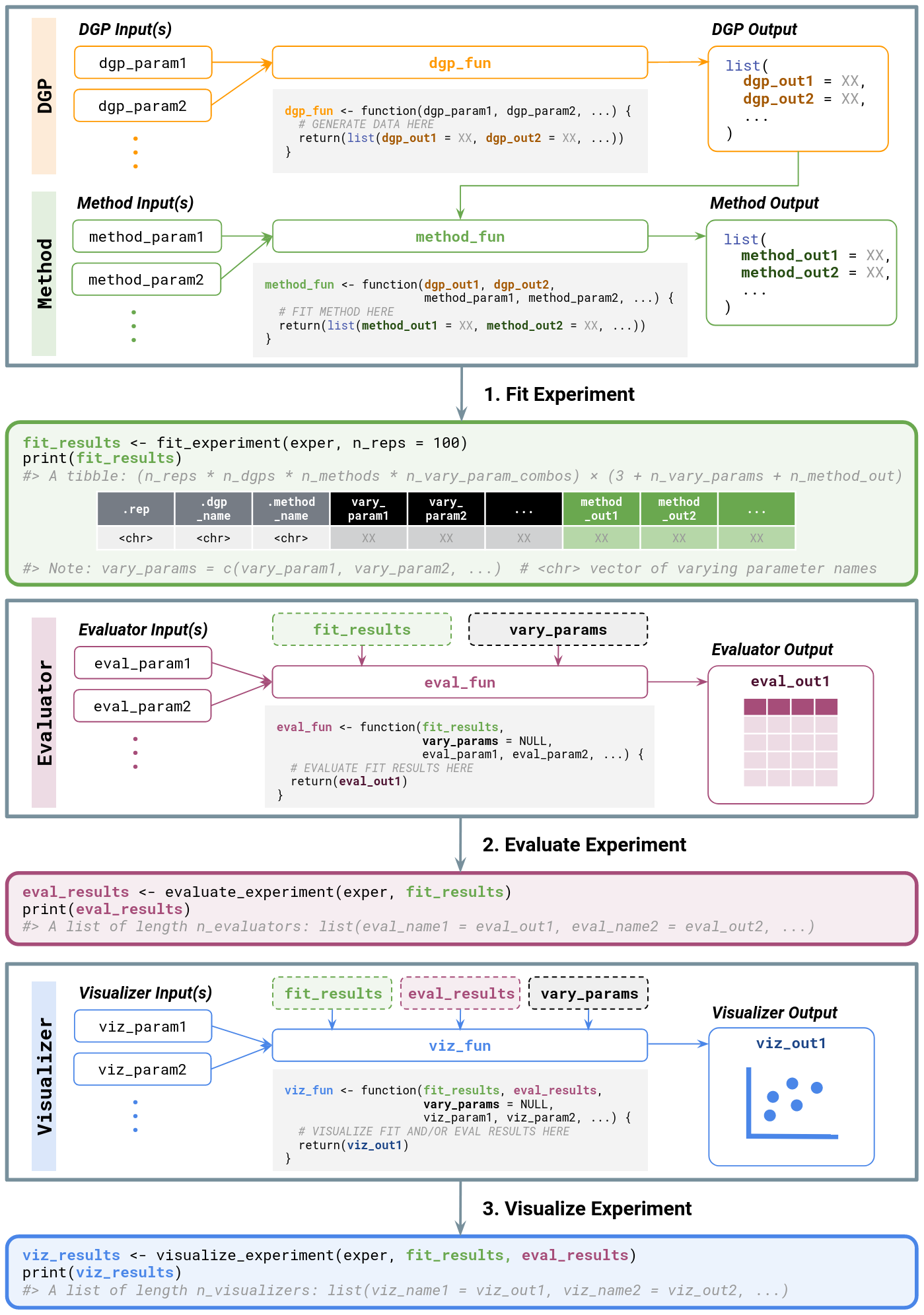
Overview of inputs and outputs for user-defined
functions when running a
simChef Experiment.
Dotted boxes denote optional input arguments that are automatically
passed through in a run_experiment() call if
specified.DGP
-
Inputs
- Any number of named parameters (e.g.,
dgp_param1,dgp_param2, …)
- Any number of named parameters (e.g.,
-
Outputs
- A list of named elements to be passed onto the method(s) (e.g.,
dgp_out1,dgp_out2, …)
- A list of named elements to be passed onto the method(s) (e.g.,
Method
-
Inputs
- Named parameters matching all names in the list returned by the DGP
function (e.g.,
dgp_out1,dgp_out2, …) - Any number of additional named parameters (e.g.,
method_param1,method_param2, …)
- Named parameters matching all names in the list returned by the DGP
function (e.g.,
-
Outputs
- A list of named elements (e.g.,
method_out1,method_out2, …)
- A list of named elements (e.g.,
Evaluator
-
Inputs
-
fit_results(optional): output offit_experiment() -
vary_params(optional): character vector of parameter names that are varied across in the experiment - Any number of additional named parameters (e.g.,
eval_param1,eval_param2, …)
-
-
Outputs
- Typically a
tibbleordata.frame
- Typically a
Visualizer
-
Inputs
-
fit_results(optional): output offit_experiment() -
eval_results(optional): output ofevaluate_experiment() -
vary_params(optional): character vector of parameter names that are varied across in the experiment - Any number of additional named parameters (e.g.,
viz_param1,viz_param2, …)
-
-
Outputs
- Typically a plot (e.g., a
ggplotorplotlyobject). Can also return tables or more generally, any R Markdown snippet (e.g., verbatim text)
- Typically a plot (e.g., a
Outputs of *_experiment
-
fit_results: output offit_experiment()- A tibble of results from each (replicate, DGP, Method) combination
with the following columns:
-
.rep: replicate ID -
.dgp_name: name of DGP -
.method_name: name of Method -
vary_acrossparameter columns (if applicable): value of the specifiedvary_acrossparameter - A column corresponding to each named component in the list returned
by
Method(e.g.,method_out1,method_out2, …)
-
- A tibble of results from each (replicate, DGP, Method) combination
with the following columns:
-
eval_results: output ofevaluate_experiment()- A named list of elements, with one element for each
Evaluatorin theExperiment- The list names correspond to the names given to each
Evaluatorincreate_evaluator(.name = ...) - The list elements are exactly the return values from each
Evaluatorfunction
- The list names correspond to the names given to each
- A named list of elements, with one element for each
-
viz_results: output ofvisualize_experiment()- A named list of elements, with one element for each
Visualizerin theExperiment- The list names correspond to the names given to each
Visualizerincreate_visualizer(.name = ...) - The list elements are exactly the return values from each
Visualizerfunction
- The list names correspond to the names given to each
- A named list of elements, with one element for each